In response to questions & comments by @hippopedoid, @adamgayoso, @akshaykagrawal et al. on "The Specious Art of Single-Cell Genomics", Tara Chari & I have posted an update with some new results. Tl;dr: definitely time to stop making t-SNE & UMAP plots.🧵biorxiv.org/content/10.110…
There's a new preprint calling to stop using UMAP, t-SNE et al, calling them arbitrary "Specious Art", which I disagree with. It's getting a lot of attention, and I think I have an idea why.
I am late to the party (was on holidays), but have now read @lpachter's "Specious Art" paper as well as ~300 quote tweets/threads, played with the code, and can add my two cents. Spoiler: I disagree with their conclusions. Some claims re t-SNE/UMAP are misleading. Thread. 🐘 pic.twitter.com/Ky9QRmXoyS twitter.com/lpachter/statu…
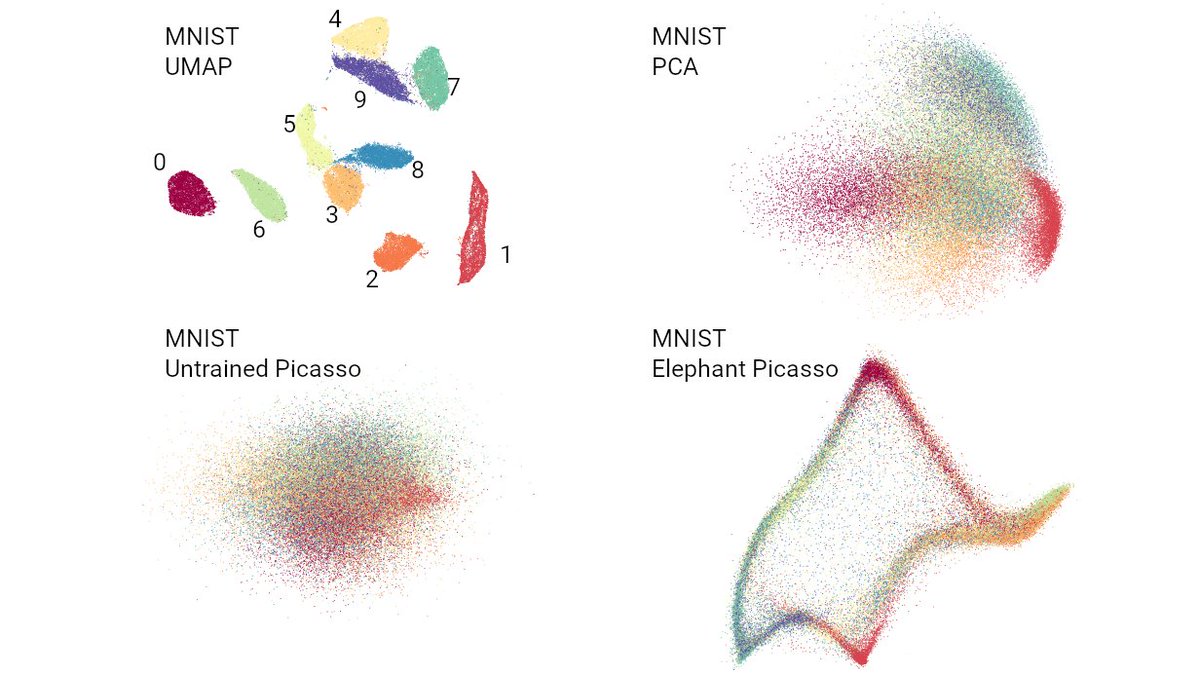
It's time to stop making t-SNE & UMAP plots. In a new preprint w/ Tara Chari we show that while they display some correlation with the underlying high-dimension data, they don't preserve local or global structure & are misleading. They're also arbitrary.🧵biorxiv.org/content/10.110… pic.twitter.com/dmFzD5RR6R
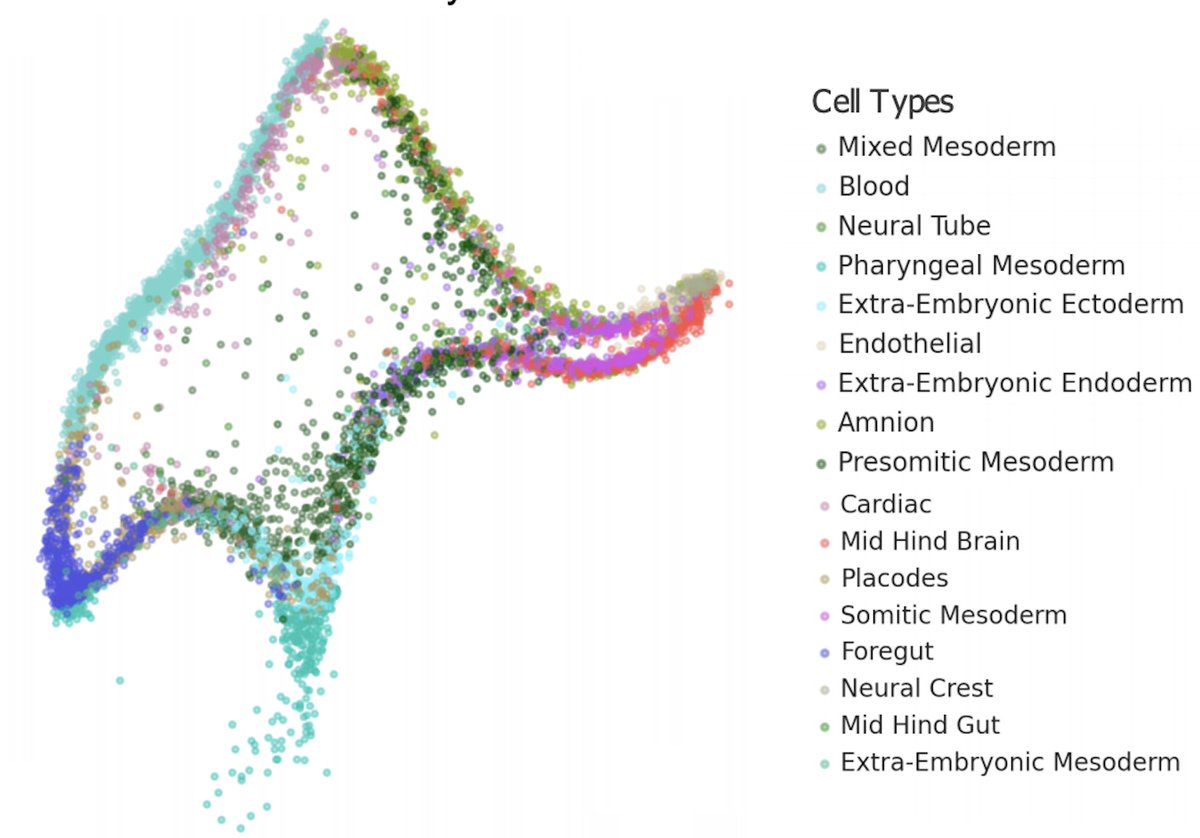
new preprint, a collaboration with @EricJDeeds, @ShamusCooley84, and @Timothy54253493. Goal: find the true dimensionality of a cell using single cell RNA-Seq. TL;DR: dimensionality reduction distorts >95% of local structure, calling into question many scRNA-Seq analyses. 1/17
what are the current methods to determine the optimal number of clusters from a single-cell RNAseq dataset?

